Jonathon C.O. Mifsud
PhD Student – Virus Evolution – Bioinformatics
Hey, I'm Jon
I am a Postdoc in the Evolutionary and Computational Virology Lab under Philippe Lemey at KU Leuven, Belgium.
Broadly, my research focuses on reconstructing the evolutionary history of viruses using phylogenetics and metagenomic sequence data.
My current work and research interests include:
- Viral protein structural phylogenetics
- Cophylogenetics and virus-host evolution
- Virus discovery and classification
Feel free to get in touch if you are after collaboration in any of these areas.
Selected Publications
-
-
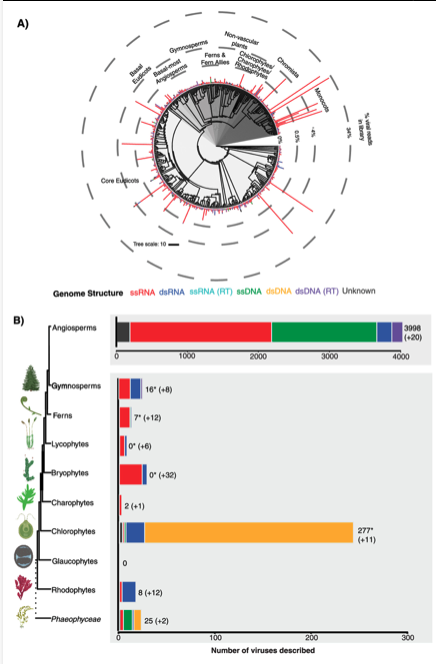 Transcriptome Mining Expands Knowledge of RNA Viruses across the Plant KingdomJournal of Virology, Dec 2022
Transcriptome Mining Expands Knowledge of RNA Viruses across the Plant KingdomJournal of Virology, Dec 2022 -
 Slippery when wet: cross-species transmission of divergent coronaviruses in bony and jawless fish and the evolutionary history of the CoronaviridaeVirus Evolution, May 2021
Slippery when wet: cross-species transmission of divergent coronaviruses in bony and jawless fish and the evolutionary history of the CoronaviridaeVirus Evolution, May 2021